DNA Cleared Plasma
- Ideal control reagent for cfDNA studies
- Better mimics patient samples than artificial matrices
- Possesses the same stability as artificial matrices
- Free of known communicable diseases (HIV, HBV, HCV, HTLV, Syphilis).
- Ready-to-use
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| DCP100A-1 | DNA Cleared Plasma | 100 mL | $557 |
|
||||
Overview
Ensure quality and accuracy of cfDNA assays
As increasing numbers of studies use liquid biopsies to discover and profile biomarkers based on cell free DNA (cfDNA), researchers need reagents that can help ensure the accuracy and precision of their cfDNA detection, quantitation, and isolation protocols, reagents like DNA Cleared Plasma.
DNA Cleared Plasma is plasma from healthy human donors that is depleted of any detectable free DNA (Figure 1) and has an otherwise similar biochemical and physiological composition as untreated plasma (Table 1, Figures 2, 3). It is an ideal blank matrix for assay development, validation, and quality control (QC) studies, such as limit of detection (LoD) determination, and spiked-in DNA remains stable for at least 270 days at 2-8°C (Figure 4). DNA Cleared Plasma better replicates the biochemical and physical characteristics of patient samples (Figure 5).
- Ideal control reagent for cfDNA studies
- Better mimics patient samples than artificial matrices
- Possesses the same stability as artificial matrices
- Free of known communicable diseases (HIV, HBV, HCV, HTLV, Syphilis).
- Ready-to-use
References
How It Works
Supporting Data
Free DNA is undetectable in DNA Cleared Plasma
Bioanalyzer analysis demonstrates the absence of detectable free DNA in DNA Cleared Plasma, making it a suitable blank baseline control that is ideal for liquid biopsy studies (Figure 1).
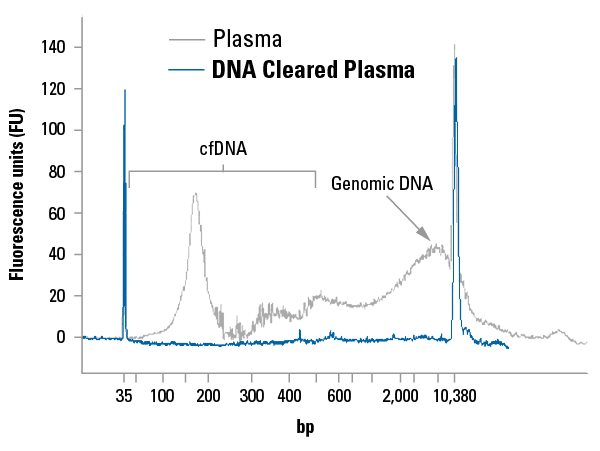
Figure 1. DNA Cleared Plasma is free of detectable cfDNA and carried-over genomic DNA.
DNA Cleared Plasma has a similar composition to untreated plasma
Analysis of DNA Cleared Plasma shows that it is compositionally similar to natural human plasma (Table 1) with comparable amounts of extracellular vesicles (EVs) (Figure 2) that are morphologically normal (Figure 3).
Table 1. DNA Cleared Plasma is biochemically similar to untreated plasma
| DNA | <0.07 pg/mL |
|---|---|
| Total protein | 5 - 7 g/dL |
| Immunoglobulin G | 500 - 1700 mg/dL |
| Immunoglobulin A | 90 - 400 mg/dL |
| Immunoglobulin M | 20 - 172 mg/dL |
| Total cholesterol | 50 - 199 mg/dL |
| Triglycerides | 50 - 149 mg/dL |
| HDL cholesterol | > 20 mg/dL |
| VLDL cholesterol | 10 - 40 mg/dL |
| LDL cholesterol | 20 - 99 mg/dL |
| pH | 7.2 - 7.6 |
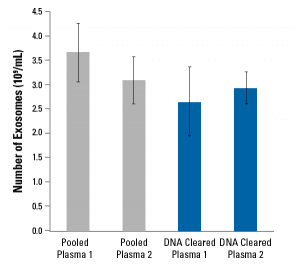
Figure 2. DNA Cleared Plasma has similar abundance of EVs as untreated plasma as determined using SBI’s EXOCET Exosome Quantitation Kit
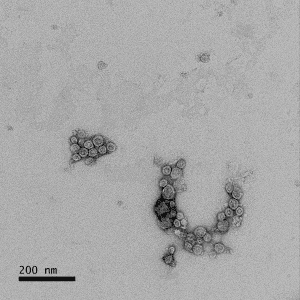
Figure 3. Transmission electron microscopy (TEM) of DNA Cleared Plasma shows the presence and unaltered morphology of EVs.
DNA Cleared Plasma is better than synthetic plasma for spike-in studies
To demonstrate the excellent stability of spiked-in reference DNA in DNA Cleared Plasma, we added either a synthetic DNA fragment (blue circles) or fragmented nucleosomal DNA (yellow squares) to DNA Cleared Plasma for up to 270 days at 2-8°C (Figure 4). At 0, 2, 4, 90, 180, and 270 days, we extracted the spiked-in DNA and used qPCR to measure the number of copies of the fragments. Similar amounts of recovery was observed at all time points.
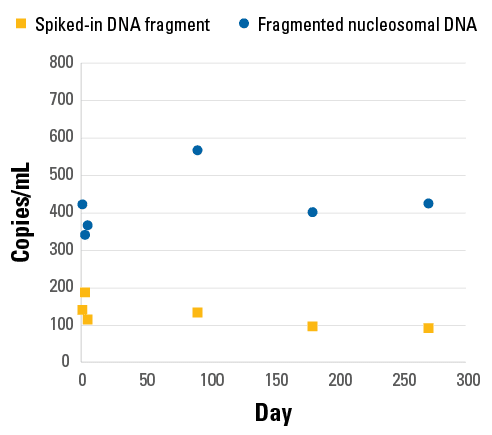
Figure 4. Reference spiked-in DNA remains stable in DNA Cleared Plasma for at least 270 days at 2-8°C.
DNA Cleared Plasma also better replicates the DNA extraction properties of natural human plasma than either synthetic plasma or buffer (Figure 5). We spiked-in a known amount of a synthetic DNA fragment into either buffer (light gray diamonds), synthetic plasma (medium gray squares), untreated plasma (dark gray triangles) or DNA Cleared Plasma (blue circles), and then extracted the DNA and compared the observed versus the expected number of copies/mL, as measured by qPCR. Of the different matrices, the DNA Cleared Plasma behaved most similarly to untreated plasma, demonstrating its suitability as a control matrix for cfDNA studies.
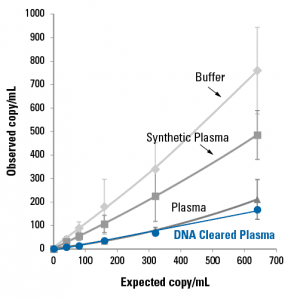
Figure 5. DNA Cleared Plasma better replicates untreated plasma than buffer or synthetic plasma for DNA extraction studies.
DNA Cleared Plasma is an excellent matrix for comparing different cfDNA extraction kits
A known amount of reference DNA was spiked into DNA Cleared Plasma and used as the starting material for a study comparing the extraction efficiency (Figure 6) and sensitivity (Figure 7) of different cfDNA isolation kits. The amount of recovered cfDNA was evaluated using qPCR, enabling quantitative comparison of the different cfDNA isolation kits. Thus, DNA Cleared Plasma can be used as a validation tool for different cfDNA isolation kits and/or cfDNA isolation method development.
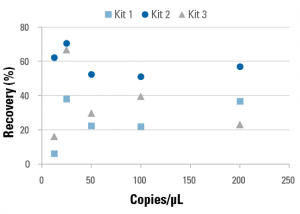
Figure 6. DNA Cleared Plasma simplifies cfDNA kit comparisons.
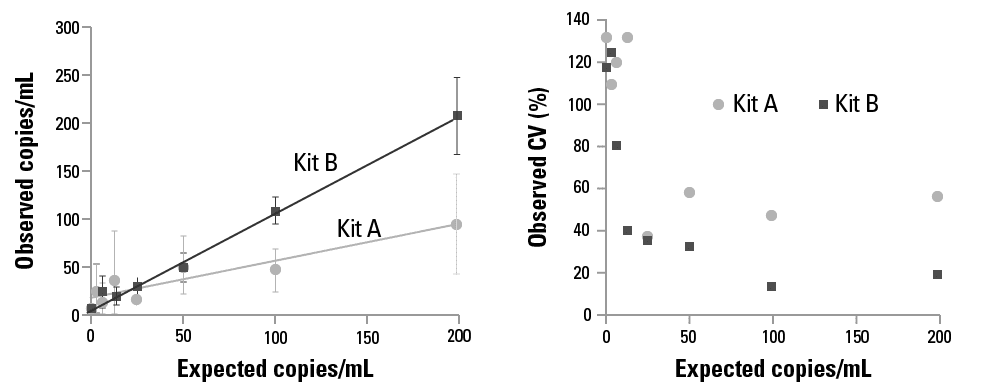
Figure 7. DNA Cleared Plasma enables comparison of limit of detection between kits.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| DCP100A-1 | DNA Cleared Plasma | 100 mL | $557 |
|
||||
Overview
Ensure quality and accuracy of cfDNA assays
As increasing numbers of studies use liquid biopsies to discover and profile biomarkers based on cell free DNA (cfDNA), researchers need reagents that can help ensure the accuracy and precision of their cfDNA detection, quantitation, and isolation protocols, reagents like DNA Cleared Plasma.
DNA Cleared Plasma is plasma from healthy human donors that is depleted of any detectable free DNA (Figure 1) and has an otherwise similar biochemical and physiological composition as untreated plasma (Table 1, Figures 2, 3). It is an ideal blank matrix for assay development, validation, and quality control (QC) studies, such as limit of detection (LoD) determination, and spiked-in DNA remains stable for at least 270 days at 2-8°C (Figure 4). DNA Cleared Plasma better replicates the biochemical and physical characteristics of patient samples (Figure 5).
- Ideal control reagent for cfDNA studies
- Better mimics patient samples than artificial matrices
- Possesses the same stability as artificial matrices
- Free of known communicable diseases (HIV, HBV, HCV, HTLV, Syphilis).
- Ready-to-use
References
How It Works
Supporting Data
Free DNA is undetectable in DNA Cleared Plasma
Bioanalyzer analysis demonstrates the absence of detectable free DNA in DNA Cleared Plasma, making it a suitable blank baseline control that is ideal for liquid biopsy studies (Figure 1).

Figure 1. DNA Cleared Plasma is free of detectable cfDNA and carried-over genomic DNA.
DNA Cleared Plasma has a similar composition to untreated plasma
Analysis of DNA Cleared Plasma shows that it is compositionally similar to natural human plasma (Table 1) with comparable amounts of extracellular vesicles (EVs) (Figure 2) that are morphologically normal (Figure 3).
Table 1. DNA Cleared Plasma is biochemically similar to untreated plasma
| DNA | <0.07 pg/mL |
|---|---|
| Total protein | 5 - 7 g/dL |
| Immunoglobulin G | 500 - 1700 mg/dL |
| Immunoglobulin A | 90 - 400 mg/dL |
| Immunoglobulin M | 20 - 172 mg/dL |
| Total cholesterol | 50 - 199 mg/dL |
| Triglycerides | 50 - 149 mg/dL |
| HDL cholesterol | > 20 mg/dL |
| VLDL cholesterol | 10 - 40 mg/dL |
| LDL cholesterol | 20 - 99 mg/dL |
| pH | 7.2 - 7.6 |

Figure 2. DNA Cleared Plasma has similar abundance of EVs as untreated plasma as determined using SBI’s EXOCET Exosome Quantitation Kit

Figure 3. Transmission electron microscopy (TEM) of DNA Cleared Plasma shows the presence and unaltered morphology of EVs.
DNA Cleared Plasma is better than synthetic plasma for spike-in studies
To demonstrate the excellent stability of spiked-in reference DNA in DNA Cleared Plasma, we added either a synthetic DNA fragment (blue circles) or fragmented nucleosomal DNA (yellow squares) to DNA Cleared Plasma for up to 270 days at 2-8°C (Figure 4). At 0, 2, 4, 90, 180, and 270 days, we extracted the spiked-in DNA and used qPCR to measure the number of copies of the fragments. Similar amounts of recovery was observed at all time points.

Figure 4. Reference spiked-in DNA remains stable in DNA Cleared Plasma for at least 270 days at 2-8°C.
DNA Cleared Plasma also better replicates the DNA extraction properties of natural human plasma than either synthetic plasma or buffer (Figure 5). We spiked-in a known amount of a synthetic DNA fragment into either buffer (light gray diamonds), synthetic plasma (medium gray squares), untreated plasma (dark gray triangles) or DNA Cleared Plasma (blue circles), and then extracted the DNA and compared the observed versus the expected number of copies/mL, as measured by qPCR. Of the different matrices, the DNA Cleared Plasma behaved most similarly to untreated plasma, demonstrating its suitability as a control matrix for cfDNA studies.

Figure 5. DNA Cleared Plasma better replicates untreated plasma than buffer or synthetic plasma for DNA extraction studies.
DNA Cleared Plasma is an excellent matrix for comparing different cfDNA extraction kits
A known amount of reference DNA was spiked into DNA Cleared Plasma and used as the starting material for a study comparing the extraction efficiency (Figure 6) and sensitivity (Figure 7) of different cfDNA isolation kits. The amount of recovered cfDNA was evaluated using qPCR, enabling quantitative comparison of the different cfDNA isolation kits. Thus, DNA Cleared Plasma can be used as a validation tool for different cfDNA isolation kits and/or cfDNA isolation method development.

Figure 6. DNA Cleared Plasma simplifies cfDNA kit comparisons.

Figure 7. DNA Cleared Plasma enables comparison of limit of detection between kits.

