
Protein Biomarker for Spinal Muscle Atrophy, Sci. Rep. 2017
Given the entire discussion above, it’s evident that extracellular vesicles can play an important role in health and disease.
Let’s take a more specific look at how this is the case with a review of some recent and interesting exosome research.
We’ll start with a study published in Scientific Reports, which showcases the importance of exosome-based protein biomarkers.
In this study, researchers took a look at the connection between spinal muscular atrophy (SMA), survival motor neuron (SMN) genes, exosomes, and survival motor neuron (SMN) protein.
SMA is a genetic disease. It’s caused by the homozygous mutation or deletion of the SMN1 gene; where the severity of the disease is then modulated by the number of copies of the highly similar SMN2 gene (Diagram 2.1).
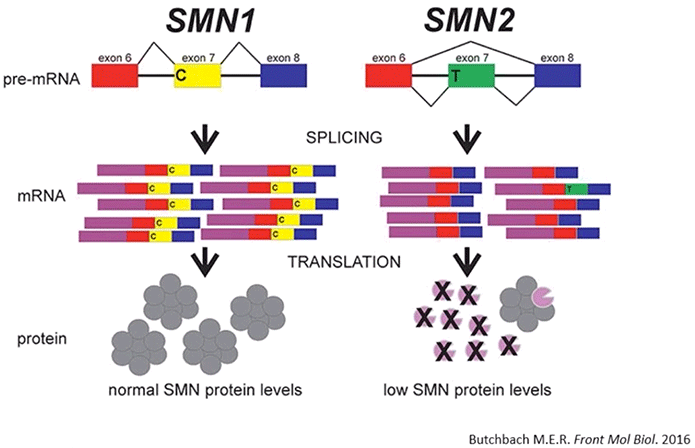
Diagram 2.1: Survival Motor Neuron Protein in SMA. At least with respect to genetic disorders, the most common cause of death in newborns is spinal muscular atrophy (SMA). SMA is characterized by the deficiency of full-length survival motor neuron (SMN) protein, the degeneration of α-motor neurons, and subsequent skeletal muscle atrophy. SMA is caused by homozygous deletion or mutation of the SMN1 gene. While people do have a highly similar copy of the SMN1 gene (SMN2), a synonymous point mutation therein results in the production of an mRNA devoid of exon 7. SMN∆7 protein is thus unstable and is unable to function like the full-length SMN protein.
The end result in SMA is that there’s a significant deficiency of full length SMN protein; and the severity of SMA is inversely correlated with the amount of SMN protein being produced.
As you may be aware, SMA is a highly debilitating neuromuscular disorder. And although antisense oligonucleotide therapy (Spinraza) was recently approved for it, there’s still a lot of room left for an objective and minimally-invasive way by which disease severity, prognosis, and even therapeutic direction can be more accurately ascertained.
This is where the measurement of exosome-derived SMN protein biomarkers comes in via this study.
The examination of protein found in TCA precipitates from numerous cell lines revealed that SMN protein is normally released from cells and into the extracellular environment in cell culture media (Diagram 2.2).
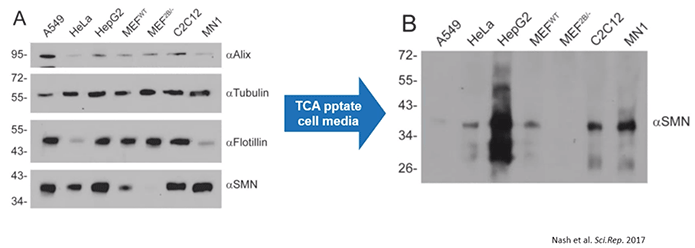
Diagram 2.2: SMN Protein is Released into the Extracellular Milieu.Cells release SMN protein into the extracellular environment. According to the study’s authors: “Various cell lines were plated in 35 mm dishes for 24 h. Media was removed, the cells lysed with protein loading buffer to analyse intracellular protein content (Panel A), and the media subjected to TCA precipitation to analyse extracellular protein content (Panel B). Equal volumes of protein sample were subjected to SDS-PAGE, transferred to a nylon membrane and probed by immunoblot with antibodies to Alix, tubulin, flotillin, or SMN. Data is representative of n = 3.
Given that, it was then important to determine if SMN protein could be detected in extracellular vesicles, especially exosomes. As a result, the authors performed electron microscopy using immunogold labeling. With this, they determined that the exosomes clearly contained SMN protein while there was very little SMN protein outside of any type of vesicle.
In brief, this means that cells naturally release SMN protein via exosomes (Diagram 2.3).
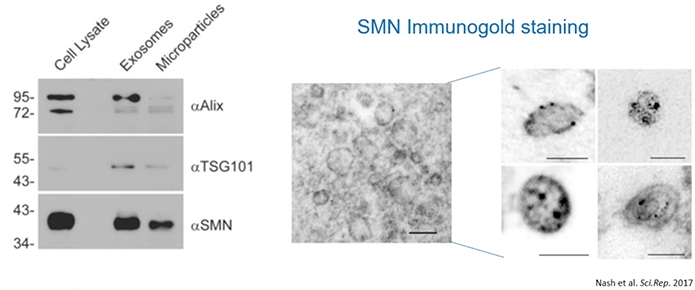
Diagram 2.3: SMN Protein Released into the Extracellular Milieu is Confined in EVs.The authors started out by isolating microparticles and EVs from “A549 cells, grown in medium supplemented with vesicle-depleted FBS, using differential centrifugation and their size profiles were determined using nanoparticle tracking analysis. Protein from extracellular vesicles (5 μg) was subjected to SDS-PAGE and the resulting immunoblot was probed for Alix, TSG101, and SMN… Immunogold labeling of SMN protein was used to demonstrate the presence of SMN protein within A549-derived exosomes (scale bar represents 100 nm)…and higher magnification of electron microscopy images showing SMN protein contained in exosomes [was performed] (scale bar represents 100 nm).”
Then, using a mouse model of SMA, the scientists determined that the quantity of SMN protein within exosomes clearly reflected the intracellular contents and disease state. Healthy mice contained high levels of SMN protein in their exosomes while affected mice had low or undetectable levels of SMN protein.
Similar results were found in healthy controls, carriers, and SMA patients. Meaning, carriers had lower levels of exosome-based SMN protein than control while those with SMA had barely detectable levels of SMN protein within exosomes (Diagram 2.4).
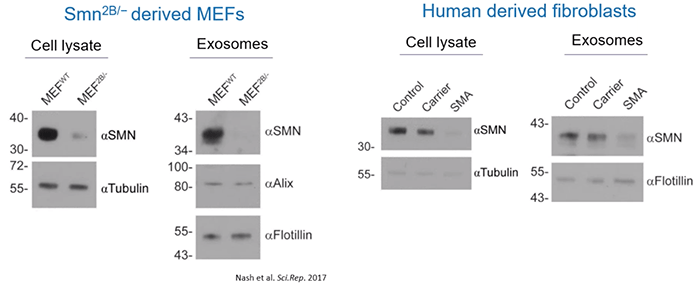
Diagram 2.4: Quantity of SMN Protein within Exosomes Correlates with Host Cell Protein Expression Levels.
Further still, the authors quantified the levels of exosomes themselves. Importantly, they observed an increased number of exosomes in the serum of patients with SMA as compared to control.
Overall, patients with SMA can be expected to have elevated levels of exosomes in their serum but lower quantities of SMN protein within those same vesicles (Diagram 2.5).
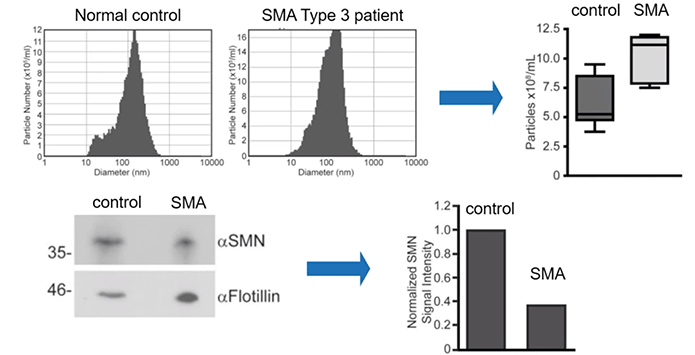
Diagram 2.5: SMN Protein Levels in Human Serum Derived EVs Reflect the Disease State.
Include same citation as 2.4
These results have important implications. They suggest that there may be a practical way by which we can diagnose and monitor SMA, including response to treatment, via the quantification of exosome-based SMN protein biomarkers and the concentration of serum-derived exosomes themselves. All of this, using minimally invasive techniques—no less.
Download the Ebook
Click here to download the pdf version of the ebook


