pCDH-EF1a-Piranha™-T2A-Puro Pre-packaged Lentivirus
- Specific—avoid off-target effects associated with genomic-based approaches (CRISPR/Cas9) or mRNA targeting (RNAi) by directly targeting the protein-of-interest
- Fast—observe protein degradation in as little as 1 hour* for speedier, more efficient studies
- Simple—relies on only two reagents, the Piranha System and your validated antibody
- Powerful—antibody-based targeting means the Piranha System can selectively degrade the phosphorylated or other post-translationally modified form of a protein while leaving the unmodified form intact
- Scalable for screening—use either the HEK293T Piranha Cell Line or your own cell line stably expressing Piranha protein to evaluate multiple protein targets in parallel (96- or 384-well plate assays)
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PTPD527VA-1 | pCDH-EF1a-Piranha™-T2A-Puro Lentivirus | >1x10^6 IFUs | $930 |
|
||||
Overview
Overview
Control protein activity quickly and specifically
When you’d like to create your own Piranha™ Targeted Protein Degradation-expressing cell line, SBI’s exclusive, ready-to-transduce pCDH-EF1α-Piranha™-T2A-Puro Pre-pacakged Lentivirus, provides an option for moderate expression in most cell types, including primary cells and stem cells. This construct is marked with puromycin for robust and easy selection of positive transductants. To initiate targeted protein degradation in your Piranha-expressing cell line, simply electroporate in antibody specific for your protein-of-interest—you’ll start seeing degradation of the protein-of-interest in as little as 1 hour*.
While studying protein function in vivo often involves knocking down expression using siRNA or knocking out the gene via CRISPR/Cas9, these approaches are indirect and often imperfect. siRNA can reduce or eliminate protein expression, but if the protein turn-over rate is slow then it can take days to see a reduction in function. Simply knocking out the gene coding for the protein-of-interest is not always possible, limits the type of studies you can do, and is incompatible with essential genes. To give researchers another faster option, SBI developed the Piranha Targeted Protein Degradation System.
- Specific—avoid off-target effects associated with genomic-based approaches (CRISPR/Cas9) or mRNA targeting (RNAi) by directly targeting the protein-of-interest
- Fast—observe protein degradation in as little as 1 hour* for speedier, more efficient studies
- Simple—relies on only two reagents, the Piranha System and your validated antibody
- Powerful—antibody-based targeting means the Piranha System can selectively degrade the phosphorylated or other post-translationally modified form of a protein while leaving the unmodified form intact
- Scalable for screening—use either the HEK293T Piranha Cell Line or your own cell line stably expressing Piranha protein to evaluate multiple protein targets in parallel (96- or 384-well plate assays)
- Unique—the only commercially available system for direct targeted degradation of protein(s)-of-interest for phenotypic studies
- Flexible—available as electroporation-ready mRNA, constitutively expressed in a HEK293T cell line, or as pre-packaged lentivirus for stable Piranha expression in the cell line of your choice
*Based on published data by Clift et al. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell. 2017 Dec 14;171(7):1692-1706.e18. doi: 10.1016/j.cell.2017.10.033. Degradation rate of target protein and resulting phenotypic effects will depend on the specific protein being targeted and influenced by protein degradation pathway and any compensatory mechanisms. An initial time course experiment to find optimal conditions to assay for protein knockdown and phenotypes is highly recommended.
Available Piranha Targeted Protein Degradation System Products
Table 1. Available Piranha Targeted Protein Degradation System Products
| Cat. # | Product |
|---|---|
| Electroporation-ready mRNA | |
| PTPD500A-1 | Piranha Electroporation-ready untagged mRNA |
| PTPD510A-1 | Piranha Electroporation-ready RFP-tagged mRNA |
| PTPD520A-1 | Piranha Electroporation-ready GFP-tagged mRNA |
| Stable Cell Line | |
| PTPD600A-1 | Piranha HEK293T GFP/Puromycin Stable Cell Line |
| Pre-packaged Lentivirus | |
| PTPD513VA-1 | pCDH-CMV-Piranha-EF1-GFP-T2A-Puro Pre-packaged Lentivirus |
| PTPD527VA-1 | pCDH-EF1a-Piranha-T2A-Puro Pre-packaged Lentivirus |
References
How It Works
How It Works
Targeted Protein Degradation with Piranha
SBI’s Piranha Targeted Protein Degradation System is based on the TRIM21 pathway found in most mammalian cells1 (Figure 1). Like TRIM21, the Piranha protein binds to conserved regions of an antibody, targeting the antibody—and the bound antigen—for degradation by the proteasome.
To use your newly-created Piranha-expressing cell line for targeted protein degradation, simply electroporate a validated antibody specific for the protein-of-interest into target cells, wait an appropriate amount of time, and then observe the phenotypic outcome. Because the rate of protein degradation is based upon many factors, we recommend performing initial experiments to empirically determine the degradation rate of your target protein and the optimal time for phenotypic observation.
1. Clift D, et al. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell. 2017 Dec 14;171(7):1692-1706.e18. doi: 10.1016/j.cell.2017.10.033.
Supporting Data
Supporting Data
See the Piranha Targeted Protein Degradation System in action
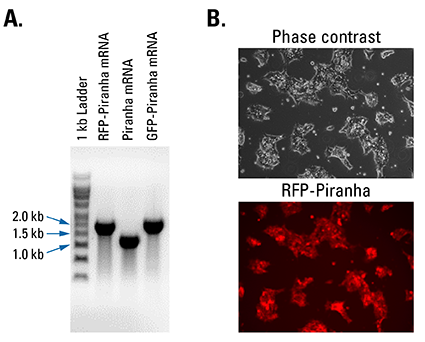
Figure 2. Piranha Electroporation-ready mRNA is pure and efficiently delivered and translated inside cells. (A) Denaturing RNA gel electrophoresis of Piranha RFP-tagged, untagged, and GFP-tagged mRNA shows clean transcripts of an appropriate size. (B) Fluorescence imaging of HEK293T cells electroporated with Piranha RFP-tagged mRNA shows robust delivery and translation of the construct.
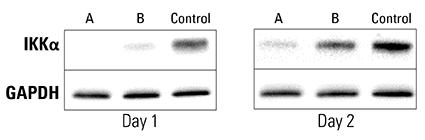
Figure 3. Piranha HEK293T Cells electroporated with anti-IKKα antibody show complete and near-complete ablation of IKKα after one day. A validated anti-IKKα antibody was electroporated into Piranha HEK293T cells using two different conditions (A and B), and IKKα protein levels assessed via Western blot and compared to a no-construct control. On day 1 post-electroporation, condition A shows complete ablation of IKKα protein and condition B shows almost complete ablation. By Day 2 levels of IKKα protein begin to return to pre-electroporation levels as the amount of antibody dissipates.
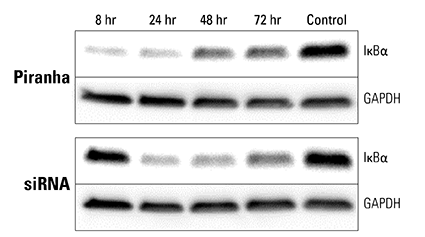
Figure 4. Piranha mRNA and antibody efficiently reduce protein levels at a faster rate than corresponding siRNA. Piranha mRNA and validated anti-IκBα antibody were co-electroporated into HeLa cells (top panel) and IκBα protein levels monitored over time via Western blot. Compared to IκBα levels after introduction of IκBα siRNA (bottom panel), the Piranha System reduces IκBα protein levels faster—reduction is apparent by 8 hours (top panel), whereas for siRNA protein reduction is first apparent only after 24 hours (bottom panel).
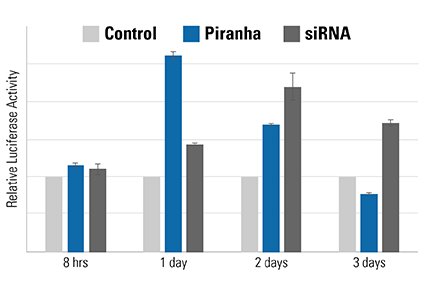
Figure 5. The speed of the Piranha System is also evident using a luciferase-based activity assay. MDA-MB-231 breast cancer cells were co-electroporated with Piranha mRNA and anti-IκBα antibody, and IκBα activity measured using an NFκB luciferase reporter assay—lower IκBα levels result in higher NFκB luciferase reporter activity. Compared to reduction of IκBα activity by siRNA (dark gray bars), which peaks on day 2, the Piranha System shows peak reduction of IκBα activity on day 1 (blue bars). The negative control is shown as light gray bars.
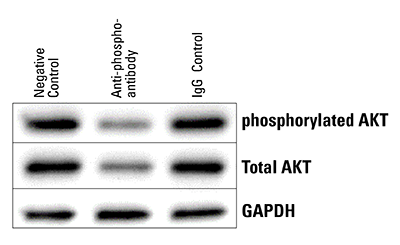
Figure 6. The Piranha System can reduce levels of phosphorylated protein. Piranha HEK293T Cells were electroporated with either buffer only (NC), anti-phospho-AKT(S473) antibody, or an IgG control, and total AKT levels assessed via Western blot. Within 5-hrs post-electroporation, both phosphorylated and total AKT protein levels are reduced.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| PTPD527VA-1 | pCDH-EF1a-Piranha™-T2A-Puro Lentivirus | >1x10^6 IFUs | $930 |
|
||||
Overview
Overview
Control protein activity quickly and specifically
When you’d like to create your own Piranha™ Targeted Protein Degradation-expressing cell line, SBI’s exclusive, ready-to-transduce pCDH-EF1α-Piranha™-T2A-Puro Pre-pacakged Lentivirus, provides an option for moderate expression in most cell types, including primary cells and stem cells. This construct is marked with puromycin for robust and easy selection of positive transductants. To initiate targeted protein degradation in your Piranha-expressing cell line, simply electroporate in antibody specific for your protein-of-interest—you’ll start seeing degradation of the protein-of-interest in as little as 1 hour*.
While studying protein function in vivo often involves knocking down expression using siRNA or knocking out the gene via CRISPR/Cas9, these approaches are indirect and often imperfect. siRNA can reduce or eliminate protein expression, but if the protein turn-over rate is slow then it can take days to see a reduction in function. Simply knocking out the gene coding for the protein-of-interest is not always possible, limits the type of studies you can do, and is incompatible with essential genes. To give researchers another faster option, SBI developed the Piranha Targeted Protein Degradation System.
- Specific—avoid off-target effects associated with genomic-based approaches (CRISPR/Cas9) or mRNA targeting (RNAi) by directly targeting the protein-of-interest
- Fast—observe protein degradation in as little as 1 hour* for speedier, more efficient studies
- Simple—relies on only two reagents, the Piranha System and your validated antibody
- Powerful—antibody-based targeting means the Piranha System can selectively degrade the phosphorylated or other post-translationally modified form of a protein while leaving the unmodified form intact
- Scalable for screening—use either the HEK293T Piranha Cell Line or your own cell line stably expressing Piranha protein to evaluate multiple protein targets in parallel (96- or 384-well plate assays)
- Unique—the only commercially available system for direct targeted degradation of protein(s)-of-interest for phenotypic studies
- Flexible—available as electroporation-ready mRNA, constitutively expressed in a HEK293T cell line, or as pre-packaged lentivirus for stable Piranha expression in the cell line of your choice
*Based on published data by Clift et al. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell. 2017 Dec 14;171(7):1692-1706.e18. doi: 10.1016/j.cell.2017.10.033. Degradation rate of target protein and resulting phenotypic effects will depend on the specific protein being targeted and influenced by protein degradation pathway and any compensatory mechanisms. An initial time course experiment to find optimal conditions to assay for protein knockdown and phenotypes is highly recommended.
Available Piranha Targeted Protein Degradation System Products
Table 1. Available Piranha Targeted Protein Degradation System Products
| Cat. # | Product |
|---|---|
| Electroporation-ready mRNA | |
| PTPD500A-1 | Piranha Electroporation-ready untagged mRNA |
| PTPD510A-1 | Piranha Electroporation-ready RFP-tagged mRNA |
| PTPD520A-1 | Piranha Electroporation-ready GFP-tagged mRNA |
| Stable Cell Line | |
| PTPD600A-1 | Piranha HEK293T GFP/Puromycin Stable Cell Line |
| Pre-packaged Lentivirus | |
| PTPD513VA-1 | pCDH-CMV-Piranha-EF1-GFP-T2A-Puro Pre-packaged Lentivirus |
| PTPD527VA-1 | pCDH-EF1a-Piranha-T2A-Puro Pre-packaged Lentivirus |
References
How It Works
How It Works
Targeted Protein Degradation with Piranha
SBI’s Piranha Targeted Protein Degradation System is based on the TRIM21 pathway found in most mammalian cells1 (Figure 1). Like TRIM21, the Piranha protein binds to conserved regions of an antibody, targeting the antibody—and the bound antigen—for degradation by the proteasome.
To use your newly-created Piranha-expressing cell line for targeted protein degradation, simply electroporate a validated antibody specific for the protein-of-interest into target cells, wait an appropriate amount of time, and then observe the phenotypic outcome. Because the rate of protein degradation is based upon many factors, we recommend performing initial experiments to empirically determine the degradation rate of your target protein and the optimal time for phenotypic observation.
1. Clift D, et al. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell. 2017 Dec 14;171(7):1692-1706.e18. doi: 10.1016/j.cell.2017.10.033.
Supporting Data
Supporting Data
See the Piranha Targeted Protein Degradation System in action
Figure 2. Piranha Electroporation-ready mRNA is pure and efficiently delivered and translated inside cells. (A) Denaturing RNA gel electrophoresis of Piranha RFP-tagged, untagged, and GFP-tagged mRNA shows clean transcripts of an appropriate size. (B) Fluorescence imaging of HEK293T cells electroporated with Piranha RFP-tagged mRNA shows robust delivery and translation of the construct.
Figure 3. Piranha HEK293T Cells electroporated with anti-IKKα antibody show complete and near-complete ablation of IKKα after one day. A validated anti-IKKα antibody was electroporated into Piranha HEK293T cells using two different conditions (A and B), and IKKα protein levels assessed via Western blot and compared to a no-construct control. On day 1 post-electroporation, condition A shows complete ablation of IKKα protein and condition B shows almost complete ablation. By Day 2 levels of IKKα protein begin to return to pre-electroporation levels as the amount of antibody dissipates.
Figure 4. Piranha mRNA and antibody efficiently reduce protein levels at a faster rate than corresponding siRNA. Piranha mRNA and validated anti-IκBα antibody were co-electroporated into HeLa cells (top panel) and IκBα protein levels monitored over time via Western blot. Compared to IκBα levels after introduction of IκBα siRNA (bottom panel), the Piranha System reduces IκBα protein levels faster—reduction is apparent by 8 hours (top panel), whereas for siRNA protein reduction is first apparent only after 24 hours (bottom panel).
Figure 5. The speed of the Piranha System is also evident using a luciferase-based activity assay. MDA-MB-231 breast cancer cells were co-electroporated with Piranha mRNA and anti-IκBα antibody, and IκBα activity measured using an NFκB luciferase reporter assay—lower IκBα levels result in higher NFκB luciferase reporter activity. Compared to reduction of IκBα activity by siRNA (dark gray bars), which peaks on day 2, the Piranha System shows peak reduction of IκBα activity on day 1 (blue bars). The negative control is shown as light gray bars.
Figure 6. The Piranha System can reduce levels of phosphorylated protein. Piranha HEK293T Cells were electroporated with either buffer only (NC), anti-phospho-AKT(S473) antibody, or an IgG control, and total AKT levels assessed via Western blot. Within 5-hrs post-electroporation, both phosphorylated and total AKT protein levels are reduced.