T7 gRNA SmartNuclease™ Cloning and Production Vector
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| CAS510A-1 | Linearized T7 gRNA SmartNuclease vector | 10 Reactions | $669 |
|
||||
Overview
Overview
Efficient gRNA cloning and production vector
The ligation-ready T7 gRNA Cloning and Production Vector is designed for efficient generation of any gRNA (also available as a complete T7 gRNA SmartNuclease Cloning and Production Kit, which includes the T7 gRNA Cloning and Production Vector as well as all the reagents you need for in vitro transcription).

References
How It Works
How It Works
Synthesizing gRNA
The workflow at-a-glance
- Design two DNA oligonucleotides that are sense and antisense sequences of the target DNA and are immediately upstream of a PAM sequence (5’ – NGG – 3’)
- Anneal the two oligonucleotides to generate a duplex
- Ligate the duplex into the pre-linearized T7 gRNA SmartNuclease Cloning and Production Vector
- Transform into competent cells and grow in LB/Kanamycin plate (50 µg/ml)
- Confirm positive clones by direct sequencing
- Perform in vitro transcription (IVT) with the T7 SmartNuclease™ gRNA Synthesis Kit (Cat.# CAS510A-KIT)
Selecting Target DNA Sequences
The selection of the target DNA sequence is not limited by any constraints, with exception of the requirement of a PAM sequence in the form of 5’ – NGG – 3’ (where N = any base) immediately following the target sequence. The typical length of the target sequence is 20 bp.
Genome Engineering
For more general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor plasmid determines whether the homologous recombination event results in a knock-out, knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Supporting Data
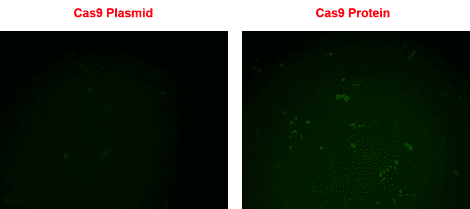
The T7 gRNA SmartNuclease Cloning and Production Vector in action
Figure 1. gRNA cloned-into and synthesized using the T7 gRNA SmartNuclease Cloning and Production Vector directs rescue of a non-fluorescent eGFP mutant using Purified Cas9 Protein. Rescue of non-fluorescent eGFP mutant (EGIP) via homology-directed repair using either a Cas9 All-in-one plasmid system (left panel) or a Cas9 protein-gRNA system (right panel), three days post-transfection. Direct transfection of Cas9 protein-gRNA results in a higher rescue efficiency.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| CAS510A-1 | Linearized T7 gRNA SmartNuclease vector | 10 Reactions | $669 |
|
||||
Overview
Overview
Efficient gRNA cloning and production vector
The ligation-ready T7 gRNA Cloning and Production Vector is designed for efficient generation of any gRNA (also available as a complete T7 gRNA SmartNuclease Cloning and Production Kit, which includes the T7 gRNA Cloning and Production Vector as well as all the reagents you need for in vitro transcription).

References
How It Works
How It Works
Synthesizing gRNA
The workflow at-a-glance
- Design two DNA oligonucleotides that are sense and antisense sequences of the target DNA and are immediately upstream of a PAM sequence (5’ – NGG – 3’)
- Anneal the two oligonucleotides to generate a duplex
- Ligate the duplex into the pre-linearized T7 gRNA SmartNuclease Cloning and Production Vector
- Transform into competent cells and grow in LB/Kanamycin plate (50 µg/ml)
- Confirm positive clones by direct sequencing
- Perform in vitro transcription (IVT) with the T7 SmartNuclease™ gRNA Synthesis Kit (Cat.# CAS510A-KIT)
Selecting Target DNA Sequences
The selection of the target DNA sequence is not limited by any constraints, with exception of the requirement of a PAM sequence in the form of 5’ – NGG – 3’ (where N = any base) immediately following the target sequence. The typical length of the target sequence is 20 bp.
Genome Engineering
For more general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor plasmid determines whether the homologous recombination event results in a knock-out, knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Supporting Data
The T7 gRNA SmartNuclease Cloning and Production Vector in action
Figure 1. gRNA cloned-into and synthesized using the T7 gRNA SmartNuclease Cloning and Production Vector directs rescue of a non-fluorescent eGFP mutant using Purified Cas9 Protein. Rescue of non-fluorescent eGFP mutant (EGIP) via homology-directed repair using either a Cas9 All-in-one plasmid system (left panel) or a Cas9 protein-gRNA system (right panel), three days post-transfection. Direct transfection of Cas9 protein-gRNA results in a higher rescue efficiency.