Exo-Flow™ 2.0 CD81 Analysis Kit for Tissue Culture Media
- Powerful—analyze EV subpopulations for deeper insights into EV biology and biomarker discovery
- Efficient—our Exo-Flow 2.0 beads can capture even small subpopulations of EVs to maximize your discoveries
- Low-background—Exo-Flow 2.0 beads deliver undetectable background binding for targeted analysis of specific EV subpopulations
- Easy-to-use—the magnetic bead-based workflow translates into quick and easy EV capture
- Willms E, et al. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol. 2018; 9: 738. PMCID: PMC5936763.
Products
Overview
Overview
Take your EV insights to the next level by focusing on specific subpopulations
“One of the biggest challenges for the EV field at this stage is addressing heterogeneity of secreted EVs, and heterogeneity within EV populations.”1Gain powerful insights into extracellular vesicle (EV) biology and analyze EV subpopulations for therapeutics development, biomarker discovery, and more with SBI’s Exo-Flow™ 2.0 CD81 Analysis Kit for Tissue Culture Media. With Exo-Flow 2.0 technology, we’ve re-engineered our original Exo-Flow beads (Figure 1) to deliver virtually undetectable background binding. The result is highly specific capture of EVs expressing CD81 on their surface so you can easily analyze specific EV subpopulations by flow cytometry, western blot, or the downstream technology of your choice. Take your EV insights to the next level with Exo-Flow 2.0.
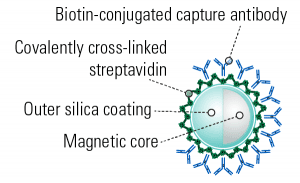
Figure 1. Exo-Flow 2.0 beads have an outer silica coating which delivers undetectable background binding.
“The new Exo-Flow 2.0 beads are much easier to see during the processing steps (sample capturing, washing, staining, etc.) which prevents accidental loss of the sample. I am also able to acquire more events with the flow cytometer with the new Exo-Flow 2.0 beads.”
—Ananthu Pucha, DeKalb Veterans Affairs Hospital at Emory University
- Powerful—analyze EV subpopulations for deeper insights into EV biology and biomarker discovery
- Efficient—our Exo-Flow 2.0 beads can capture even small subpopulations of EVs to maximize your discoveries
- Low-background—Exo-Flow 2.0 beads deliver undetectable background binding for targeted analysis of specific EV subpopulations
- Easy-to-use—the magnetic bead-based workflow translates into quick and easy EV capture
Each Exo-Flow 2.0 CD81 Analysis Kit for Tissue Culture Media comes with enough reagents for 10 reactions of 150 – 300 µg protein equivalent of EVs (the actual amount of input protein equivalent depends on your EV isolation method).
Our Exo-Flow 2.0 technology is available as both individual tetraspanin (CD63, CD9, and CD81) and combination kits, as well as a basic kit that can be used with the biotinylated antibody of your choice. Note that unlike the original Exo-Flow technology, Exo-Flow 2.0 kits are specific for isolating EVs from either serum/plasma or tissue culture medium, so be sure to choose the right kit for your needs.
See all available Exo-Flow 2.0 kits in the table below.
| Biotinylated Antibody | Exo-Flow 2.0 Kit for EV Isolation from Serum or Plasma Cat.# | Exo-Flow 2.0 Kit for EV Isolation from Tissue Culture Medium Cat.# |
|---|---|---|
| CD63 | EXOFLOW2-100A-SP | EXOFLOW2-200A-TC |
| CD9 | EXOFLOW2-105A-SP | EXOFLOW2-205A-TC |
| CD81 | EXOFLOW2-110A-SP | EXOFLOW2-210A-TC |
| Tetraspanin Combo (CD63, CD9, and CD81) | EXOFLOW2-150A-SP | EXOFLOW2-250A-TC |
| Basic Kit without antibody | EXOFLOW2-BASICA-SP | EXOFLOW2-BASICA-TC |
- Willms E, et al. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol. 2018; 9: 738. PMCID: PMC5936763.
References
How It Works
How It Works
Easily analyze EV subpopulations with Exo-Flow 2.0
The Exo-Flow 2.0 workflow uses a simple, four-step workflow:- Couple the biotinylated CD81 antibody to the magnetic streptavidin Exo-Flow 2.0 beads
- Use the anti-CD81-coupled magnetic beads to capture already-isolated EVs (compatible with ExoQuick®-TC, ultracentrifugation, or any other EV isolation method)
- Wash away unbound EVs
- Prep for analysis
- For flow cytometry—stain with the included Red or Green Dye
- For western blotting—resuspend beads in M-PER/RIPA lysis buffer (not included), add sample loading buffer load onto gel
- For RNA analysis—resuspend beads in RNA lysis buffer of your choice and process
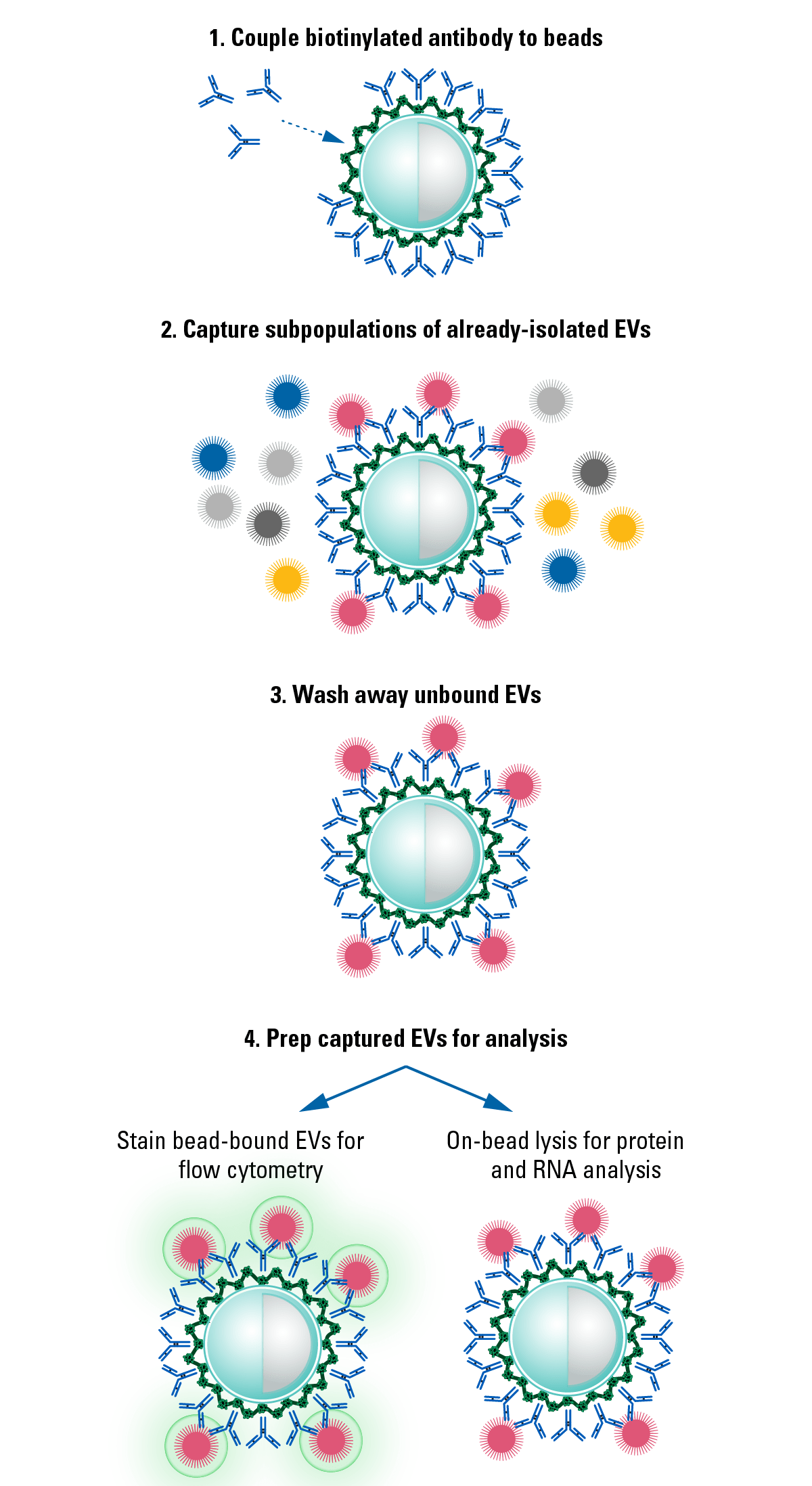
Figure 2. The simple Exo-Flow 2.0 Workflow.
Supporting Data
Supporting Data
See the performance that Exo-Flow 2.0 technology delivers
Exo-Flow 2.0 delivers undetectable background binding when analyzed by western blot
We isolated EVs from HEK293 cells and serum using ExoQuick®-TC and ExoQuick® Exosome Isolation Reagents, captured specific EV subpopulations using Exo-Flow 2.0 beads coated with either anti-CD63, anti-CD9, anti-CD81, or a no-antibody control, and analyzed the captured EVs by western blot probing for the exosome-specific marker TSG101 (Figure 3). The absence of detectable signal in the no-antibody control lane demonstrates the low background binding of Exo-Flow 2.0 technology. The varying signal intensity seen in Figure 3 also highlights the heterogeneity of EV subpopulations.
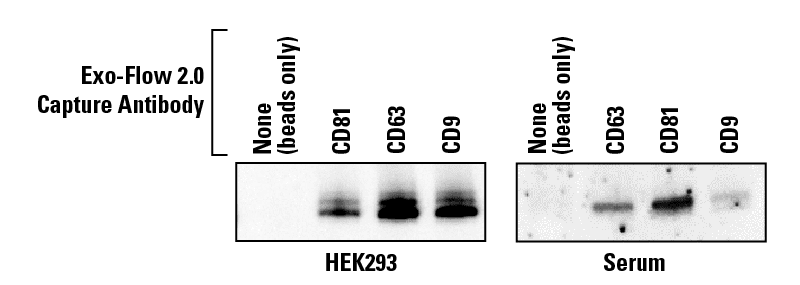
Figure 3. Exo-Flow 2.0 delivers undetectable background binding when analyzed by western blot.
Exo-Flow 2.0 delivers undetectable background binding when analyzed using flow cytometry
We isolated EVs from HEK293 cells using ExoQuick-TC and captured the subpopulation of EVs expressing CD9 (Figure 4A, dark red trace) and the subpopulation of EVs bearing CD63 (Figure 4A orange trace). We also isolated EVs from serum using the SmartSEC HT EV Isolation system and captured the subpopulation of EVs bearing CD81 (Figure 4B). After staining bead-bound EVs and washing away excess stain, we analyzed the EV subpopulations by flow cytometry. In both plots, distinct subpopulations of EVs can be seen—in Figure 4A, the peak seen on the red trace indicates a subpopulation of EVs bearing CD9 and the separate peak seen on the orange trace indicates a subpopulation of EVs bearing CD63. In Figure 4B, the peak seen on the green trace indicates a subpopulation of EVs bearing CD81.
In both plots, the overlap of signal in the beads-only and no-antibody traces demonstrates the low background binding of Exo-Flow 2.0 technology.
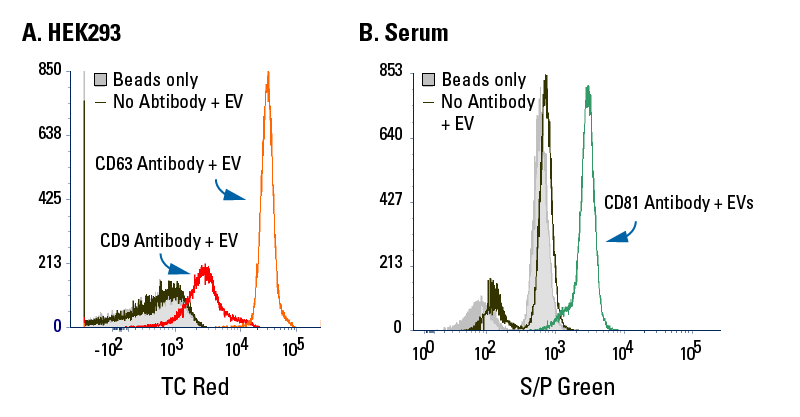
Figure 4. Exo-Flow 2.0 delivers undetectable background binding when analyzed using flow cytometry.
Understand surface marker differences between EV subpopulations with Exo-Flow 2.0
We isolated EVs from serum using SmartSEC HT, captured subpopulations of EVs using Exo-Flow 2.0 beads coupled with either biotinylated CD9 (Figure 5A) or CD63 (Figure 5B) antibodies, and analyzed these subpopulations for the presence of CD14, a myeloid lineage marker, using flow cytometry (Figure 5). The data show that EVs captured using anti-CD9 Exo-Flow 2.0 beads are more likely to also bear CD14 than EVs captured using anti-CD63- Exo-Flow 2.0 beads, demonstrating the effectiveness of Exo-Flow 2.0 technology for EV subpopulation analysis and enabling better biomarker screening.
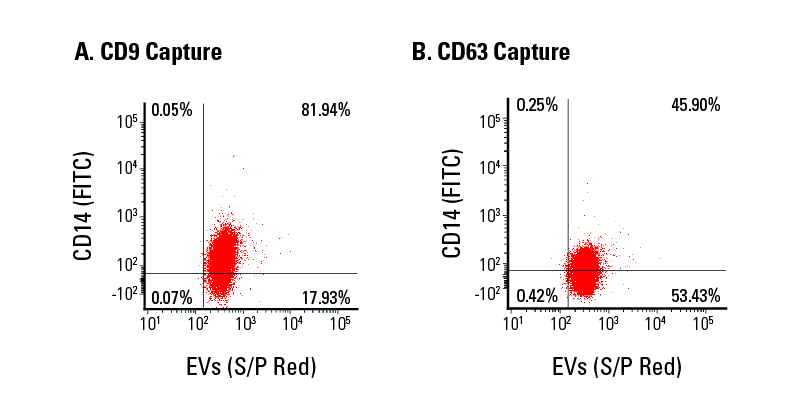
Figure 5. Exo-Flow 2.0 supports flow cytometric analysis of EV subpopulations.
Use Exo-Flow 2.0 to understand differences in cargo, such as miRNA levels, between EV subpopulations
We isolated EVs from HEK293-conditioned medium using ExoQuick-TC, captured subpopulations of EVs using Exo-Flow 2.0 beads coupled with either biotinylated CD63 or CD81 antibodies, and analyzed these subpopulations for the presence of miR-122 (Figure 6). The data show a statistically significant difference in the amount of miR-122 present in these EV subpopulations, with ~3.6-fold more miR-122 found in CD81 EVs than CD63 EVs, demonstrating the effectiveness of Exo-Flow 2.0 technology for EV subpopulation analysis.
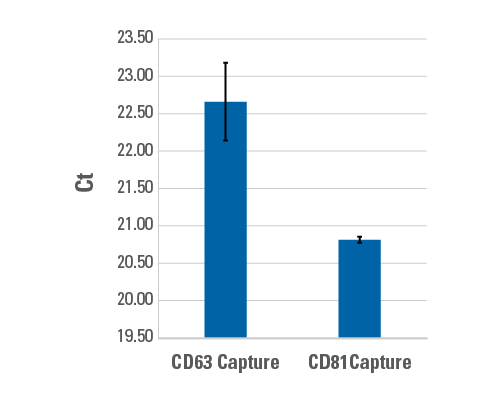
Figure 6. Use Exo-Flow 2.0 to understand differences in cargo, such as miRNA levels, between EV subpopulations.
FAQs
Documentation
Citations
Related Products
Products
Overview
Overview
Take your EV insights to the next level by focusing on specific subpopulations
“One of the biggest challenges for the EV field at this stage is addressing heterogeneity of secreted EVs, and heterogeneity within EV populations.”1Gain powerful insights into extracellular vesicle (EV) biology and analyze EV subpopulations for therapeutics development, biomarker discovery, and more with SBI’s Exo-Flow™ 2.0 CD81 Analysis Kit for Tissue Culture Media. With Exo-Flow 2.0 technology, we’ve re-engineered our original Exo-Flow beads (Figure 1) to deliver virtually undetectable background binding. The result is highly specific capture of EVs expressing CD81 on their surface so you can easily analyze specific EV subpopulations by flow cytometry, western blot, or the downstream technology of your choice. Take your EV insights to the next level with Exo-Flow 2.0.

Figure 1. Exo-Flow 2.0 beads have an outer silica coating which delivers undetectable background binding.
“The new Exo-Flow 2.0 beads are much easier to see during the processing steps (sample capturing, washing, staining, etc.) which prevents accidental loss of the sample. I am also able to acquire more events with the flow cytometer with the new Exo-Flow 2.0 beads.”
—Ananthu Pucha, DeKalb Veterans Affairs Hospital at Emory University
- Powerful—analyze EV subpopulations for deeper insights into EV biology and biomarker discovery
- Efficient—our Exo-Flow 2.0 beads can capture even small subpopulations of EVs to maximize your discoveries
- Low-background—Exo-Flow 2.0 beads deliver undetectable background binding for targeted analysis of specific EV subpopulations
- Easy-to-use—the magnetic bead-based workflow translates into quick and easy EV capture
Each Exo-Flow 2.0 CD81 Analysis Kit for Tissue Culture Media comes with enough reagents for 10 reactions of 150 – 300 µg protein equivalent of EVs (the actual amount of input protein equivalent depends on your EV isolation method).
Our Exo-Flow 2.0 technology is available as both individual tetraspanin (CD63, CD9, and CD81) and combination kits, as well as a basic kit that can be used with the biotinylated antibody of your choice. Note that unlike the original Exo-Flow technology, Exo-Flow 2.0 kits are specific for isolating EVs from either serum/plasma or tissue culture medium, so be sure to choose the right kit for your needs.
See all available Exo-Flow 2.0 kits in the table below.
| Biotinylated Antibody | Exo-Flow 2.0 Kit for EV Isolation from Serum or Plasma Cat.# | Exo-Flow 2.0 Kit for EV Isolation from Tissue Culture Medium Cat.# |
|---|---|---|
| CD63 | EXOFLOW2-100A-SP | EXOFLOW2-200A-TC |
| CD9 | EXOFLOW2-105A-SP | EXOFLOW2-205A-TC |
| CD81 | EXOFLOW2-110A-SP | EXOFLOW2-210A-TC |
| Tetraspanin Combo (CD63, CD9, and CD81) | EXOFLOW2-150A-SP | EXOFLOW2-250A-TC |
| Basic Kit without antibody | EXOFLOW2-BASICA-SP | EXOFLOW2-BASICA-TC |
- Willms E, et al. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol. 2018; 9: 738. PMCID: PMC5936763.
References
How It Works
How It Works
Easily analyze EV subpopulations with Exo-Flow 2.0
The Exo-Flow 2.0 workflow uses a simple, four-step workflow:- Couple the biotinylated CD81 antibody to the magnetic streptavidin Exo-Flow 2.0 beads
- Use the anti-CD81-coupled magnetic beads to capture already-isolated EVs (compatible with ExoQuick®-TC, ultracentrifugation, or any other EV isolation method)
- Wash away unbound EVs
- Prep for analysis
- For flow cytometry—stain with the included Red or Green Dye
- For western blotting—resuspend beads in M-PER/RIPA lysis buffer (not included), add sample loading buffer load onto gel
- For RNA analysis—resuspend beads in RNA lysis buffer of your choice and process

Figure 2. The simple Exo-Flow 2.0 Workflow.
Supporting Data
Supporting Data
See the performance that Exo-Flow 2.0 technology delivers
Exo-Flow 2.0 delivers undetectable background binding when analyzed by western blot
We isolated EVs from HEK293 cells and serum using ExoQuick®-TC and ExoQuick® Exosome Isolation Reagents, captured specific EV subpopulations using Exo-Flow 2.0 beads coated with either anti-CD63, anti-CD9, anti-CD81, or a no-antibody control, and analyzed the captured EVs by western blot probing for the exosome-specific marker TSG101 (Figure 3). The absence of detectable signal in the no-antibody control lane demonstrates the low background binding of Exo-Flow 2.0 technology. The varying signal intensity seen in Figure 3 also highlights the heterogeneity of EV subpopulations.

Figure 3. Exo-Flow 2.0 delivers undetectable background binding when analyzed by western blot.
Exo-Flow 2.0 delivers undetectable background binding when analyzed using flow cytometry
We isolated EVs from HEK293 cells using ExoQuick-TC and captured the subpopulation of EVs expressing CD9 (Figure 4A, dark red trace) and the subpopulation of EVs bearing CD63 (Figure 4A orange trace). We also isolated EVs from serum using the SmartSEC HT EV Isolation system and captured the subpopulation of EVs bearing CD81 (Figure 4B). After staining bead-bound EVs and washing away excess stain, we analyzed the EV subpopulations by flow cytometry. In both plots, distinct subpopulations of EVs can be seen—in Figure 4A, the peak seen on the red trace indicates a subpopulation of EVs bearing CD9 and the separate peak seen on the orange trace indicates a subpopulation of EVs bearing CD63. In Figure 4B, the peak seen on the green trace indicates a subpopulation of EVs bearing CD81.
In both plots, the overlap of signal in the beads-only and no-antibody traces demonstrates the low background binding of Exo-Flow 2.0 technology.

Figure 4. Exo-Flow 2.0 delivers undetectable background binding when analyzed using flow cytometry.
Understand surface marker differences between EV subpopulations with Exo-Flow 2.0
We isolated EVs from serum using SmartSEC HT, captured subpopulations of EVs using Exo-Flow 2.0 beads coupled with either biotinylated CD9 (Figure 5A) or CD63 (Figure 5B) antibodies, and analyzed these subpopulations for the presence of CD14, a myeloid lineage marker, using flow cytometry (Figure 5). The data show that EVs captured using anti-CD9 Exo-Flow 2.0 beads are more likely to also bear CD14 than EVs captured using anti-CD63- Exo-Flow 2.0 beads, demonstrating the effectiveness of Exo-Flow 2.0 technology for EV subpopulation analysis and enabling better biomarker screening.

Figure 5. Exo-Flow 2.0 supports flow cytometric analysis of EV subpopulations.
Use Exo-Flow 2.0 to understand differences in cargo, such as miRNA levels, between EV subpopulations
We isolated EVs from HEK293-conditioned medium using ExoQuick-TC, captured subpopulations of EVs using Exo-Flow 2.0 beads coupled with either biotinylated CD63 or CD81 antibodies, and analyzed these subpopulations for the presence of miR-122 (Figure 6). The data show a statistically significant difference in the amount of miR-122 present in these EV subpopulations, with ~3.6-fold more miR-122 found in CD81 EVs than CD63 EVs, demonstrating the effectiveness of Exo-Flow 2.0 technology for EV subpopulation analysis.

Figure 6. Use Exo-Flow 2.0 to understand differences in cargo, such as miRNA levels, between EV subpopulations.

