Mouse pre-miRNA Scramble Negative Control Expression Lentivector
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| MMIR-000-PA-1 | Mouse pre-miRNA Scramble Negative Control Expression Lentivector | Bacterial Streak | $454 |
|
||||
Overview
Overview
Supporting your studies with ready-to-go negative controls
No need to make a negative control for your Mouse Precursor miRNA Expression Lentivector Studies—SBI’s already built one for you. With the Mouse pre-miRNA Scramble Negative Control Expression Lentivector, you get the same lentivector that expresses our targeted miRNAs, but with a scrambled, non-targeting stem-loop sequence so you can easily control for increased miRNA effects.
Why choose SBI’s precursor miRNA lentivectors?
Whether you’re using miRNAs to study cellular processes or develop the next generation of therapeutics, SBI’s comprehensive collection of mouse precursor miRNA lentivectors are a superior alternative to synthetic RNAs. Like synthetic miRs, SBI’s precursor miRNAs can be transfected into target cells for transient miR expression. But unlike synthetic miRs, they can also be transduced into a variety of target cells—including primary cells, stem cells, and other hard-to-transfect cell lines—to create stable miR-producing cell lines, maximizing your options for miR expression.
In addition, SBI’s precursor miRNA lentivectors are designed for efficient expression and accurate processing into mature miRNAs, for native miR-like behavior. Each construct is cloned with the native stem-loop structure plus an additional 200 – 400 bps of upstream and downstream DNA, ensuring that the miRNA processing machinery operates just as it would with the native transcript.

And because our collection includes cancer and stem cell-related miRNA clusters, we offer a wider range of constructs than just the mature miRNA sequences listed in miRbase.
The precursor miRNA Expression Lentivectors drive expression of the miR precursor with the constitutive CMV promoter, for strong expression in common cell types including HeLa, HEK293, and HT1080 cell lines. Transductants/transfectants can be easily selected and sorted using copGFP and puromycin markers.
- Transfect or transduce the precursor miRNA lentivectors
- Create stably-expressing miR cell lines
- Leverage SBI’s highly-regarded lentivectors
- Obtain accurate miR processing and efficient expression through the use of native precursor sequences
- Use copGFP markers to identify desired clones
How It Works
Supporting Data
Supporting Data
Get high levels of miRs from our Mouse pre-miRNA Expression Lentivectors
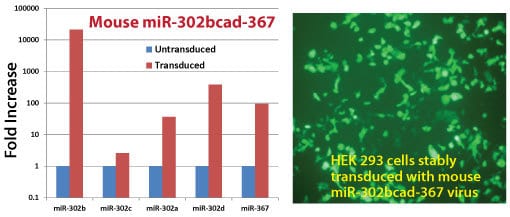
METHODS: HEK 293 cells were transduced with the mouse miR-302bcad-367 (Cat.# MMIR-302bcad+367-PA-CL) packaged lentivirus. After 48-hours, cellular RNA was analyzed by Real-time qPCR using SBI’s QuantiMir small RNA quantification system (Cat.# RA420A-1). The expression levels of miR-302-b, -c,-a,-d and -367 were measured and compared to non-transduced cellular controls. Robust mature microRNA overexpression was observed in the transduced cells when compared to controls. Cells were also monitored for GFP using fluorescent microscopy.
FAQs
Resources
Related Products
Citations
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| MMIR-000-PA-1 | Mouse pre-miRNA Scramble Negative Control Expression Lentivector | Bacterial Streak | $454 |
|
||||
Overview
Overview
Supporting your studies with ready-to-go negative controls
No need to make a negative control for your Mouse Precursor miRNA Expression Lentivector Studies—SBI’s already built one for you. With the Mouse pre-miRNA Scramble Negative Control Expression Lentivector, you get the same lentivector that expresses our targeted miRNAs, but with a scrambled, non-targeting stem-loop sequence so you can easily control for increased miRNA effects.
Why choose SBI’s precursor miRNA lentivectors?
Whether you’re using miRNAs to study cellular processes or develop the next generation of therapeutics, SBI’s comprehensive collection of mouse precursor miRNA lentivectors are a superior alternative to synthetic RNAs. Like synthetic miRs, SBI’s precursor miRNAs can be transfected into target cells for transient miR expression. But unlike synthetic miRs, they can also be transduced into a variety of target cells—including primary cells, stem cells, and other hard-to-transfect cell lines—to create stable miR-producing cell lines, maximizing your options for miR expression.
In addition, SBI’s precursor miRNA lentivectors are designed for efficient expression and accurate processing into mature miRNAs, for native miR-like behavior. Each construct is cloned with the native stem-loop structure plus an additional 200 – 400 bps of upstream and downstream DNA, ensuring that the miRNA processing machinery operates just as it would with the native transcript.

And because our collection includes cancer and stem cell-related miRNA clusters, we offer a wider range of constructs than just the mature miRNA sequences listed in miRbase.
The precursor miRNA Expression Lentivectors drive expression of the miR precursor with the constitutive CMV promoter, for strong expression in common cell types including HeLa, HEK293, and HT1080 cell lines. Transductants/transfectants can be easily selected and sorted using copGFP and puromycin markers.
- Transfect or transduce the precursor miRNA lentivectors
- Create stably-expressing miR cell lines
- Leverage SBI’s highly-regarded lentivectors
- Obtain accurate miR processing and efficient expression through the use of native precursor sequences
- Use copGFP markers to identify desired clones
Supporting Data
Supporting Data
Get high levels of miRs from our Mouse pre-miRNA Expression Lentivectors
METHODS: HEK 293 cells were transduced with the mouse miR-302bcad-367 (Cat.# MMIR-302bcad+367-PA-CL) packaged lentivirus. After 48-hours, cellular RNA was analyzed by Real-time qPCR using SBI’s QuantiMir small RNA quantification system (Cat.# RA420A-1). The expression levels of miR-302-b, -c,-a,-d and -367 were measured and compared to non-transduced cellular controls. Robust mature microRNA overexpression was observed in the transduced cells when compared to controls. Cells were also monitored for GFP using fluorescent microscopy.