CAG-T7-hspCas9-H1-gRNA All-in-one Cas9 SmartNuclease™ Plasmid
- Conveniently deliver Cas9 and gRNA with a single vector
- Drive Cas9 expression with the CAG promoter, which provides high expression levels in primary cells and stem cells
- Express gRNA from the H1 promoter for maximum specificity and choice of targets
- Ensure efficient import of Cas9 to the nucleus with N-term and C-term nuclear localization signals (NLSs)
- Boost Cas9 gene expression and stabilize the transcript via the WPRE regulatory element after the C-term NLS
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| CAS720A-1 | All-in-one CAG-T7-hspCas9-H1-gRNA linearized SmartNuclease vector | 10 Reactions | $779 |
|
||||
Overview
Overview
Simplified Cas9 and gRNA delivery
All-in-one Cas9 and gRNA plasmids are an excellent way to simplify delivery of your CRISPR/Cas9 system by providing both Cas9 and gRNA from a single vector. SBI’s CAG-T7-hspCas9-H1-gRNA All-in-one Cas9 SmartNuclease Plasmid includes a number of features that make it a great All-in-one choice for any genome engineering project involving transfectable cells:

- Conveniently deliver Cas9 and gRNA with a single vector
- Drive Cas9 expression with the CAG promoter, which provides high expression levels in primary cells and stem cells
- Express gRNA from the H1 promoter for maximum specificity and choice of targets
- Ensure efficient import of Cas9 to the nucleus with N-term and C-term nuclear localization signals (NLSs)
- Boost Cas9 gene expression and stabilize the transcript via the WPRE regulatory element after the C-term NLS
- Easily detect and/or purify the Cas9 protein with the N-term myc-tag
- Produce Cas9 mRNA via in vitro transcription using the convenient T7 promoter
As with all of our Cas9 delivery options, the CAG-T7-hspCas9-H1-gRNA Plasmid is functionally validated and comes backed by our expert technical support team—if you’ve got a genome engineering question just ask by emailing tech@systembio.com.
Why an HR targeting vector is a recommended
Even though gene knock-outs can result from DSBs caused by Cas9 alone, SBI recommends the use of HR targeting vectors (also called HR donor vectors) for more efficient and precise mutation. HR donors can supply elements for positive or negative selection ensuring easier identification of successful mutation events. In addition, HR donors can include up to 6-8 kb of open reading frame for gene knock-ins or tagging, and, when small mutations are included in either 5’ or 3’ homology arms, can make specific, targeted gene edits.
Not sure whether you need a CRISPR/Cas9 plasmid, purified protein, or mRNA?
Use this table to choose the CRISPR/Cas9 product that’s right for you:
For This Application | In these types of cells | Use These Products |
|---|---|---|
MODIFYING ORGANISMS
| Embryos—to create transgenic animals | Injectable Cas9 mRNA & gRNA Synthesis Kits Cas9 Protein EGFP-labeled Cas9 Protein |
| Animals models—in vivo genome editing | AAV-Cas9 Vectors Cas9 Protein EGFP-labeled Cas9 Protein |
|
MODIFYING CELL LINES
| Cells that are transfectable | Cas9 Plasmids Cas9 Protein EGFP-labeled Cas9 Protein |
Difficult-to-transfect cell lines:
| AAV-Cas9 Vectors Lenti Cas9 Systems |
|
SCREENING
| All cell types requiring stable Cas9 overexpression | Lenti Cas9 Systems AAVS1 Safe Harbor Site Cas9 Gene Knock-in System Cas9 Protein EGFP-labeled Cas9 Protein |
PRE-CLINICAL APPLICATIONS
| All cell types and applications | Cas9 Nickase, available in all delivery formats Cas9 Protein EGFP-labeled Cas9 Protein |
| SIMULTANEOUS ENGINEERING OF MULTIPLE MUTATIONS | All cell types and applications | Multiplex gRNA cloning kit, compatible with all Cas9 delivery options |
References
How It Works
How It Works
Using SBI’s All-in-one Cas9 SmartNuclease Plasmids
The workflow at-a-glance
- Design two DNA oligonucleotides that are sense and antisense sequences of the target DNA and are immediately upstream of a PAM sequence (5’ – NGG – 3’)
- Anneal the two oligonucleotides to generate a duplex
- Ligate the duplex into the pre-linearized All-in-one Cas9 SmartNuclease Plasmid
- Transform into competent cells and grow in LB/Kanamycin plate (50 µg/ml)
- Confirm positive clones by direct sequencing
- Transfect sequence-verified All-in-one construct into mammalian cells using standard transfection protocols (co-transfect with an HR Targeting Vector, if required for your application)
- Perform a Surveyor Nuclease assay (or other suitable mismatch cleavage assays) to check for site-specific genome cleavage and select for desired clones
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor plasmid determines whether the homologous recombination event results in a knock-out, knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Supporting Data
Genome engineering with SBI’s All-in-one Cas9 SmartNuclease Plasmids
Note that this study uses a slightly different All-in-one Cas9 SmartNuclease Plasmid design, but the results are expected to be similar.
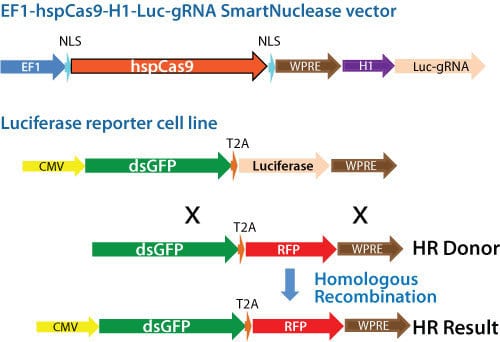
Figure 1. Schematic showing knock-out of a genomic luciferase gene by insertion of an RFP gene. Cas9 activity and gRNA were delivered using an All-in-one Cas9 SmartNuclease Plasmid.
To demonstrate the capabilities of our All-in-one Cas9 SmartNuclease Plasmids and also show how the choice of gRNA sequence can affect genome editing efficiencies, we designed a study to knock-out a genomic copy of a luciferase gene by inserting a copy of the RFP gene. This study used two different All-in-one constructs—EF1-hspCas9-H1-Luc-gRNA1 and EF1-hspCas9-H1-Luc-gRNA2—and an HR donor vector containing the RFP gene surrounded by sequences homologous to the DNA around the genomic luciferase gene (Figure 1). While both constructs enabled successful genome editing, as indicated by the recovery of RFP-positive clones, the construct with gRNA1 provided more efficient genome editing than the construct with gRNA2 (Figure 2).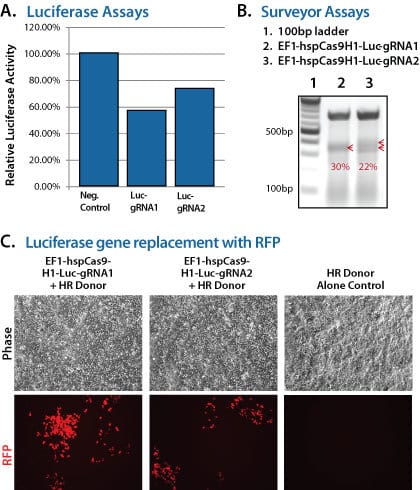
Figure 2. The All-in-one Cas9 SmartNuclease Plasmid enabled successful genomic luciferase knock-out and replacement with RFP. (A) Insertion of the RFP gene into the genomically-integrated luciferase gene results in a decrease in luciferase activity, indicating successful genome editing. (B) A Surveyor Nuclease Assay reflects finding from (A) that the gRNA1 construct delivered slightly more cleavage than the gRNA2 construct. (C) Fluorescence imaging shows successful genome editing by the recovery of RFP-positive clones.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| CAS720A-1 | All-in-one CAG-T7-hspCas9-H1-gRNA linearized SmartNuclease vector | 10 Reactions | $779 |
|
||||
Overview
Overview
Simplified Cas9 and gRNA delivery
All-in-one Cas9 and gRNA plasmids are an excellent way to simplify delivery of your CRISPR/Cas9 system by providing both Cas9 and gRNA from a single vector. SBI’s CAG-T7-hspCas9-H1-gRNA All-in-one Cas9 SmartNuclease Plasmid includes a number of features that make it a great All-in-one choice for any genome engineering project involving transfectable cells:

- Conveniently deliver Cas9 and gRNA with a single vector
- Drive Cas9 expression with the CAG promoter, which provides high expression levels in primary cells and stem cells
- Express gRNA from the H1 promoter for maximum specificity and choice of targets
- Ensure efficient import of Cas9 to the nucleus with N-term and C-term nuclear localization signals (NLSs)
- Boost Cas9 gene expression and stabilize the transcript via the WPRE regulatory element after the C-term NLS
- Easily detect and/or purify the Cas9 protein with the N-term myc-tag
- Produce Cas9 mRNA via in vitro transcription using the convenient T7 promoter
As with all of our Cas9 delivery options, the CAG-T7-hspCas9-H1-gRNA Plasmid is functionally validated and comes backed by our expert technical support team—if you’ve got a genome engineering question just ask by emailing tech@systembio.com.
Why an HR targeting vector is a recommended
Even though gene knock-outs can result from DSBs caused by Cas9 alone, SBI recommends the use of HR targeting vectors (also called HR donor vectors) for more efficient and precise mutation. HR donors can supply elements for positive or negative selection ensuring easier identification of successful mutation events. In addition, HR donors can include up to 6-8 kb of open reading frame for gene knock-ins or tagging, and, when small mutations are included in either 5’ or 3’ homology arms, can make specific, targeted gene edits.
Not sure whether you need a CRISPR/Cas9 plasmid, purified protein, or mRNA?
Use this table to choose the CRISPR/Cas9 product that’s right for you:
For This Application | In these types of cells | Use These Products |
|---|---|---|
MODIFYING ORGANISMS
| Embryos—to create transgenic animals | Injectable Cas9 mRNA & gRNA Synthesis Kits Cas9 Protein EGFP-labeled Cas9 Protein |
| Animals models—in vivo genome editing | AAV-Cas9 Vectors Cas9 Protein EGFP-labeled Cas9 Protein |
|
MODIFYING CELL LINES
| Cells that are transfectable | Cas9 Plasmids Cas9 Protein EGFP-labeled Cas9 Protein |
Difficult-to-transfect cell lines:
| AAV-Cas9 Vectors Lenti Cas9 Systems |
|
SCREENING
| All cell types requiring stable Cas9 overexpression | Lenti Cas9 Systems AAVS1 Safe Harbor Site Cas9 Gene Knock-in System Cas9 Protein EGFP-labeled Cas9 Protein |
PRE-CLINICAL APPLICATIONS
| All cell types and applications | Cas9 Nickase, available in all delivery formats Cas9 Protein EGFP-labeled Cas9 Protein |
| SIMULTANEOUS ENGINEERING OF MULTIPLE MUTATIONS | All cell types and applications | Multiplex gRNA cloning kit, compatible with all Cas9 delivery options |
References
How It Works
How It Works
Using SBI’s All-in-one Cas9 SmartNuclease Plasmids
The workflow at-a-glance
- Design two DNA oligonucleotides that are sense and antisense sequences of the target DNA and are immediately upstream of a PAM sequence (5’ – NGG – 3’)
- Anneal the two oligonucleotides to generate a duplex
- Ligate the duplex into the pre-linearized All-in-one Cas9 SmartNuclease Plasmid
- Transform into competent cells and grow in LB/Kanamycin plate (50 µg/ml)
- Confirm positive clones by direct sequencing
- Transfect sequence-verified All-in-one construct into mammalian cells using standard transfection protocols (co-transfect with an HR Targeting Vector, if required for your application)
- Perform a Surveyor Nuclease assay (or other suitable mismatch cleavage assays) to check for site-specific genome cleavage and select for desired clones
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor plasmid determines whether the homologous recombination event results in a knock-out, knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Supporting Data
Genome engineering with SBI’s All-in-one Cas9 SmartNuclease Plasmids
Note that this study uses a slightly different All-in-one Cas9 SmartNuclease Plasmid design, but the results are expected to be similar.
Figure 1. Schematic showing knock-out of a genomic luciferase gene by insertion of an RFP gene. Cas9 activity and gRNA were delivered using an All-in-one Cas9 SmartNuclease Plasmid.
To demonstrate the capabilities of our All-in-one Cas9 SmartNuclease Plasmids and also show how the choice of gRNA sequence can affect genome editing efficiencies, we designed a study to knock-out a genomic copy of a luciferase gene by inserting a copy of the RFP gene. This study used two different All-in-one constructs—EF1-hspCas9-H1-Luc-gRNA1 and EF1-hspCas9-H1-Luc-gRNA2—and an HR donor vector containing the RFP gene surrounded by sequences homologous to the DNA around the genomic luciferase gene (Figure 1). While both constructs enabled successful genome editing, as indicated by the recovery of RFP-positive clones, the construct with gRNA1 provided more efficient genome editing than the construct with gRNA2 (Figure 2).
Figure 2. The All-in-one Cas9 SmartNuclease Plasmid enabled successful genomic luciferase knock-out and replacement with RFP. (A) Insertion of the RFP gene into the genomically-integrated luciferase gene results in a decrease in luciferase activity, indicating successful genome editing. (B) A Surveyor Nuclease Assay reflects finding from (A) that the gRNA1 construct delivered slightly more cleavage than the gRNA2 construct. (C) Fluorescence imaging shows successful genome editing by the recovery of RFP-positive clones.