Cas9 SmartNickase™ mRNA, Injection- and Transfection-ready
- Ready to use for in vivo genome editing applications
- Functionally validated Cas9 SmartNickase
- Backed by expert, easy-to-reach technical support
- Ran, FA. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Oct 24; 8:2281-2308. PMCID: PMC3969860.
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| CAS504A-1 | Transfection-ready Cas9 SmartNickase mRNA (Eukaryotic Nickase mutant version) | 20 µg | $532 |
|
||||
Overview
Overview
Stay on-target in your in vivo genome editing projects
For pre-clinical and other applications where you need to minimize off-target Cas9 activity, use SBI’s injection- and transfection-ready PrecisonX™ Cas9 SmartNickase™ mRNA.
Unlike the wildtype Cas9 protein which introduces double-strand breaks (DSBs), the Cas9 SmartNickase introduces paired nicks at the gRNA-directed site. Creating nicks favors the higher-fidelity homologous recombination process over non-homologous end joining (NHEJ), with paired nicking shown to reduce off-target activity by 50- to 1,500-fold in cell lines, and to facilitate gene knockout in mice without losing on-target cleavage efficiency1.
As with all of our Cas9 delivery options, the Cas9 SmartNickase mRNA is functionally validated and comes backed by our expert technical support team—if you’ve got a genome engineering question just ask by emailing tech@systembio.com.- Ready to use for in vivo genome editing applications
- Functionally validated Cas9 SmartNickase
- Backed by expert, easy-to-reach technical support
Why an HR targeting vector is a recommended
Even though gene knock-outs can result from DSBs caused by Cas9 alone, SBI recommends the use of HR targeting vectors (also called HR donor vectors) for more efficient and precise mutation. HR donors can supply elements for positive or negative selection ensuring easier identification of successful mutation events. In addition, HR donors can include up to 6-8 kb of open reading frame for gene knock-ins or tagging, and, when small mutations are included in either 5’ or 3’ homology arms, can make specific, targeted gene edits.
Efficiently generate gRNA with the T7 gRNA SmartNickase Synthesis Kit
Pair your Cas9 SmartNickase mRNA with our T7 gRNA SmartNuclease Synthesis Kit, which comes with a pre-linearized, ready-for-cloning gRNA expression vector and all the reagents you need for T7-driven in vitro transcription of your cloned gRNA.

Not sure whether you need a CRISPR/Cas9 plasmid, purified protein, or mRNA?
Use this table to choose the CRISPR/Cas9 product that’s right for you:
For This Application | In these types of cells | Use These Products |
|---|---|---|
MODIFYING ORGANISMS
| Embryos—to create transgenic animals | Injectable Cas9 mRNA & gRNA Synthesis Kits Cas9 Protein EGFP-labeled Cas9 Protein |
| Animals models—in vivo genome editing | AAV-Cas9 Vectors Cas9 Protein EGFP-labeled Cas9 Protein |
|
MODIFYING CELL LINES
| Cells that are transfectable | Cas9 Plasmids Cas9 Protein EGFP-labeled Cas9 Protein |
Difficult-to-transfect cell lines:
| AAV-Cas9 Vectors Lenti Cas9 Systems |
|
SCREENING
| All cell types requiring stable Cas9 overexpression | Lenti Cas9 Systems AAVS1 Safe Harbor Site Cas9 Gene Knock-in System Cas9 Protein EGFP-labeled Cas9 Protein |
PRE-CLINICAL APPLICATIONS
| All cell types and applications | Cas9 Nickase, available in all delivery formats Cas9 Protein EGFP-labeled Cas9 Protein |
| SIMULTANEOUS ENGINEERING OF MULTIPLE MUTATIONS | All cell types and applications | Multiplex gRNA cloning kit, compatible with all Cas9 delivery options |
- Ran, FA. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Oct 24; 8:2281-2308. PMCID: PMC3969860.
References
How It Works
How It Works
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor plasmid determines whether the homologous recombination event results in a knock-out, knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Supporting Data
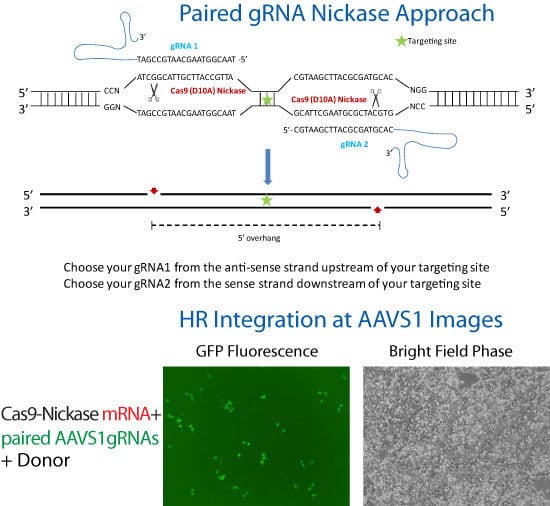
Knock-in of GFP at the AAVS1 Safe Harbor Site using Cas9 SmartNickase mRNA
Figure 1. Cas9 SmartNickase mRNA-mediated knock-in of GFP to the AAVS1 Safe Harbor Site. In order to enhance genome editing specificity, paired gRNA with the Cas9 SmartNickase can be used to generate double nicking with 5′ overhang. Please follow the guidelines in the schematic for paired gRNA selection and design—note that only gRNA pairs creating 5′ overhangs with less than 8 bp overlap between the guide sequences were able to mediate detectable indel formation1. To achieve high cleavage efficiency using Cas9 Nickase with paired gRNAs, make sure each gRNA is able to efficiently induce indels when coupled with wide-type Cas9.
References
- Ran, FA. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Oct 24; 8:2281-2308. PMCID: PMC3969860.
FAQs
Documentation
Citations
Related Products
Products
| Catalog Number | Description | Size | Price | Quantity | Add to Cart | |||
|---|---|---|---|---|---|---|---|---|
| CAS504A-1 | Transfection-ready Cas9 SmartNickase mRNA (Eukaryotic Nickase mutant version) | 20 µg | $532 |
|
||||
Overview
Overview
Stay on-target in your in vivo genome editing projects
For pre-clinical and other applications where you need to minimize off-target Cas9 activity, use SBI’s injection- and transfection-ready PrecisonX™ Cas9 SmartNickase™ mRNA.
Unlike the wildtype Cas9 protein which introduces double-strand breaks (DSBs), the Cas9 SmartNickase introduces paired nicks at the gRNA-directed site. Creating nicks favors the higher-fidelity homologous recombination process over non-homologous end joining (NHEJ), with paired nicking shown to reduce off-target activity by 50- to 1,500-fold in cell lines, and to facilitate gene knockout in mice without losing on-target cleavage efficiency1.
As with all of our Cas9 delivery options, the Cas9 SmartNickase mRNA is functionally validated and comes backed by our expert technical support team—if you’ve got a genome engineering question just ask by emailing tech@systembio.com.- Ready to use for in vivo genome editing applications
- Functionally validated Cas9 SmartNickase
- Backed by expert, easy-to-reach technical support
Why an HR targeting vector is a recommended
Even though gene knock-outs can result from DSBs caused by Cas9 alone, SBI recommends the use of HR targeting vectors (also called HR donor vectors) for more efficient and precise mutation. HR donors can supply elements for positive or negative selection ensuring easier identification of successful mutation events. In addition, HR donors can include up to 6-8 kb of open reading frame for gene knock-ins or tagging, and, when small mutations are included in either 5’ or 3’ homology arms, can make specific, targeted gene edits.
Efficiently generate gRNA with the T7 gRNA SmartNickase Synthesis Kit
Pair your Cas9 SmartNickase mRNA with our T7 gRNA SmartNuclease Synthesis Kit, which comes with a pre-linearized, ready-for-cloning gRNA expression vector and all the reagents you need for T7-driven in vitro transcription of your cloned gRNA.

Not sure whether you need a CRISPR/Cas9 plasmid, purified protein, or mRNA?
Use this table to choose the CRISPR/Cas9 product that’s right for you:
For This Application | In these types of cells | Use These Products |
|---|---|---|
MODIFYING ORGANISMS
| Embryos—to create transgenic animals | Injectable Cas9 mRNA & gRNA Synthesis Kits Cas9 Protein EGFP-labeled Cas9 Protein |
| Animals models—in vivo genome editing | AAV-Cas9 Vectors Cas9 Protein EGFP-labeled Cas9 Protein |
|
MODIFYING CELL LINES
| Cells that are transfectable | Cas9 Plasmids Cas9 Protein EGFP-labeled Cas9 Protein |
Difficult-to-transfect cell lines:
| AAV-Cas9 Vectors Lenti Cas9 Systems |
|
SCREENING
| All cell types requiring stable Cas9 overexpression | Lenti Cas9 Systems AAVS1 Safe Harbor Site Cas9 Gene Knock-in System Cas9 Protein EGFP-labeled Cas9 Protein |
PRE-CLINICAL APPLICATIONS
| All cell types and applications | Cas9 Nickase, available in all delivery formats Cas9 Protein EGFP-labeled Cas9 Protein |
| SIMULTANEOUS ENGINEERING OF MULTIPLE MUTATIONS | All cell types and applications | Multiplex gRNA cloning kit, compatible with all Cas9 delivery options |
- Ran, FA. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Oct 24; 8:2281-2308. PMCID: PMC3969860.
References
How It Works
How It Works
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor plasmid determines whether the homologous recombination event results in a knock-out, knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Supporting Data
Knock-in of GFP at the AAVS1 Safe Harbor Site using Cas9 SmartNickase mRNA
Figure 1. Cas9 SmartNickase mRNA-mediated knock-in of GFP to the AAVS1 Safe Harbor Site. In order to enhance genome editing specificity, paired gRNA with the Cas9 SmartNickase can be used to generate double nicking with 5′ overhang. Please follow the guidelines in the schematic for paired gRNA selection and design—note that only gRNA pairs creating 5′ overhangs with less than 8 bp overlap between the guide sequences were able to mediate detectable indel formation1. To achieve high cleavage efficiency using Cas9 Nickase with paired gRNAs, make sure each gRNA is able to efficiently induce indels when coupled with wide-type Cas9.
References
- Ran, FA. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Oct 24; 8:2281-2308. PMCID: PMC3969860.